A2.1.3 – EVIDENCE FOR EVOLUTION OF LIFE
📌Definition Table
| Term | Definition |
|---|---|
| LUCA | Last Universal Common Ancestor; the most recent organism from which all life on Earth descended. |
| Phylogenetic Tree | Diagram showing evolutionary relationships among species based on genetic and morphological data. |
| Molecular Clock | Method of estimating evolutionary time by measuring genetic mutations over time. |
| Hydrothermal Vent Hypothesis | Theory that life originated in deep-sea vents rich in minerals and chemical energy. |
| Radiometric Dating | Technique for determining the age of rocks or fossils by measuring isotopic ratios. |
📌Introduction
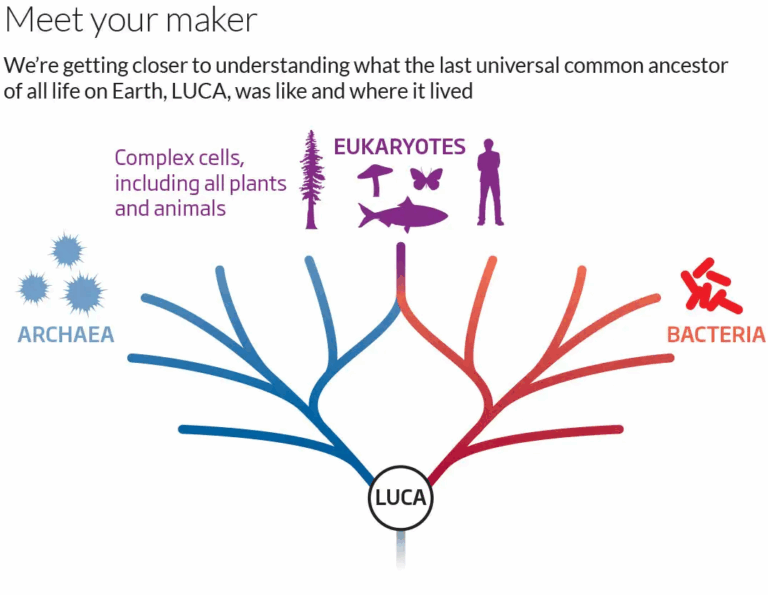
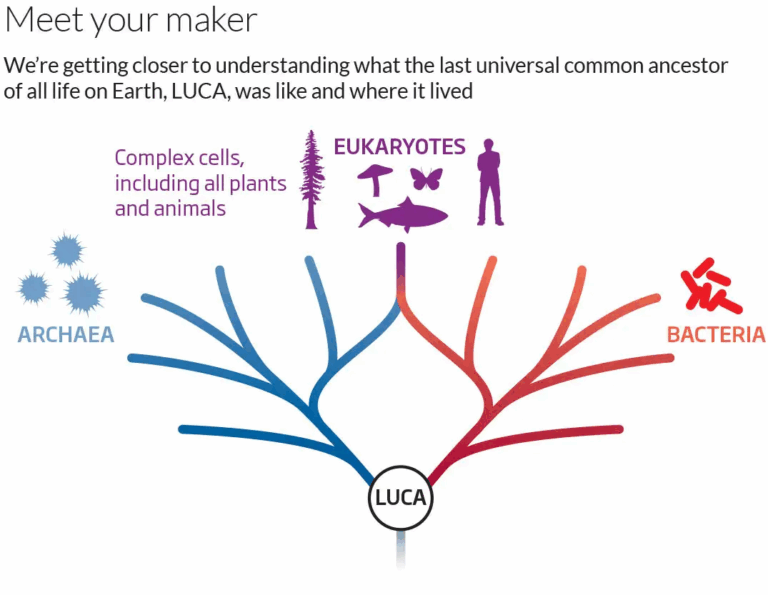
The theory that all living organisms share a common origin is supported by multiple lines of evidence, including biochemical similarities, fossil records, and genetic data. The Last Universal Common Ancestor (LUCA) is thought to have existed about 4 billion years ago, giving rise to the three major domains of life — Bacteria, Archaea, and Eukarya. Fossils, molecular clocks, and the discovery of ancient microbial structures in hydrothermal vents provide strong evidence for the evolutionary timeline. By comparing DNA sequences, amino acid structures, and metabolic pathways, scientists can reconstruct the evolutionary tree of life and trace the shared heritage of all organisms.
📌 Evidence for a Common Ancestor
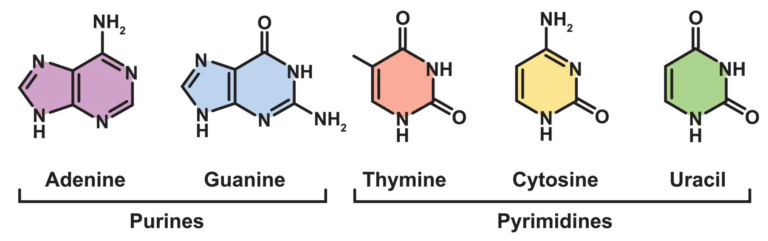
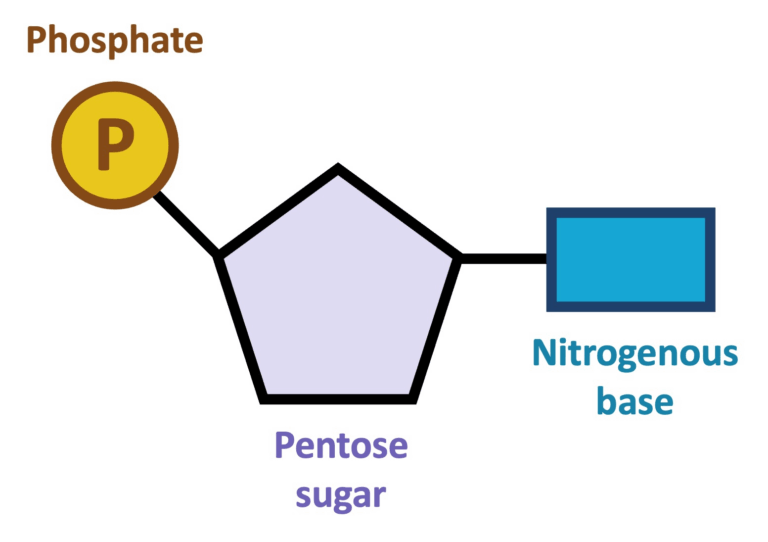
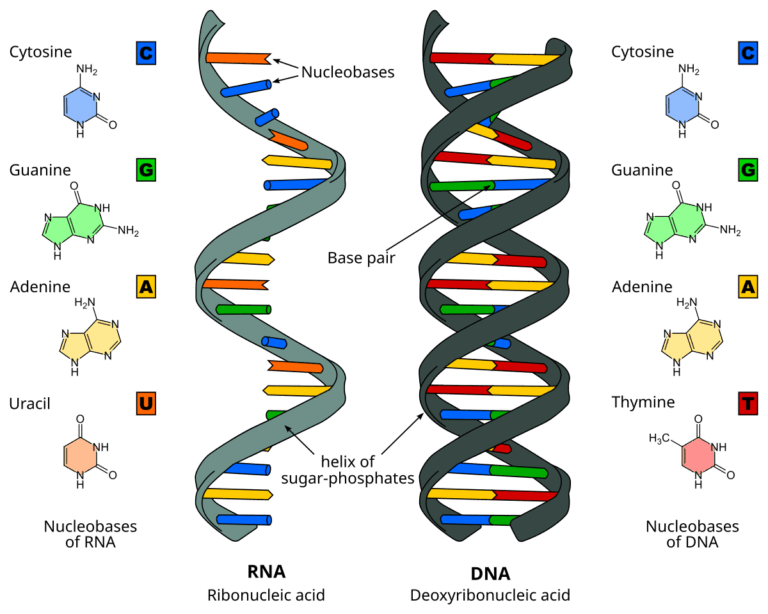
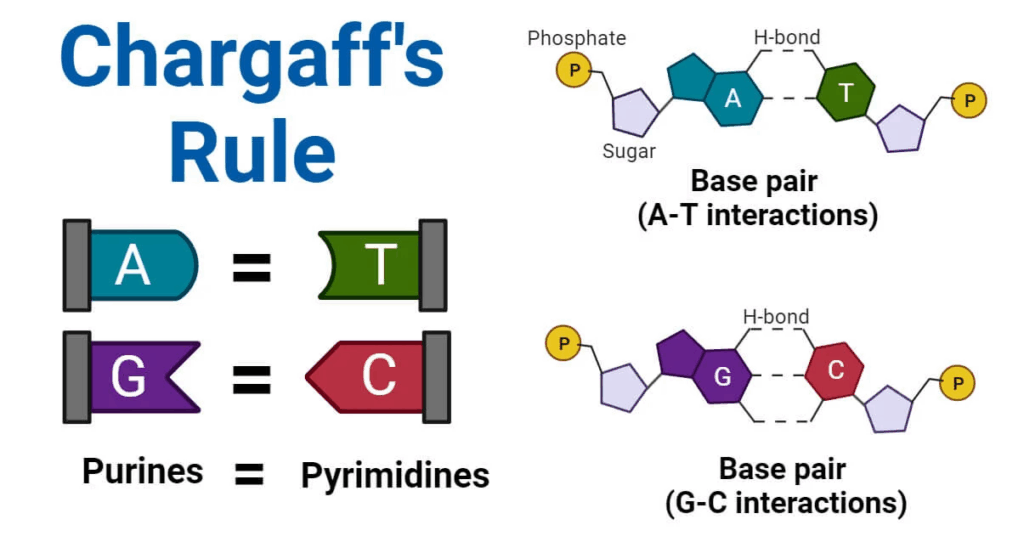
- All organisms share the same DNA bases (A, T/U, G, C) and a universal genetic code.
- The same set of amino acids is used to build proteins in all known life forms.
- Many genes are shared between Archaea and Bacteria, indicating inheritance from LUCA.
- Structural similarities, such as the vertebrate forelimb bone pattern, show morphological evidence of common descent.
- LUCA likely existed around 4 billion years ago and occupied the base of the phylogenetic tree of life.
- Other life forms existing alongside LUCA may have gone extinct due to competition.
🧠 Examiner Tip: Use both molecular and morphological evidence when explaining common ancestry — this strengthens your argument in long-answer questions.
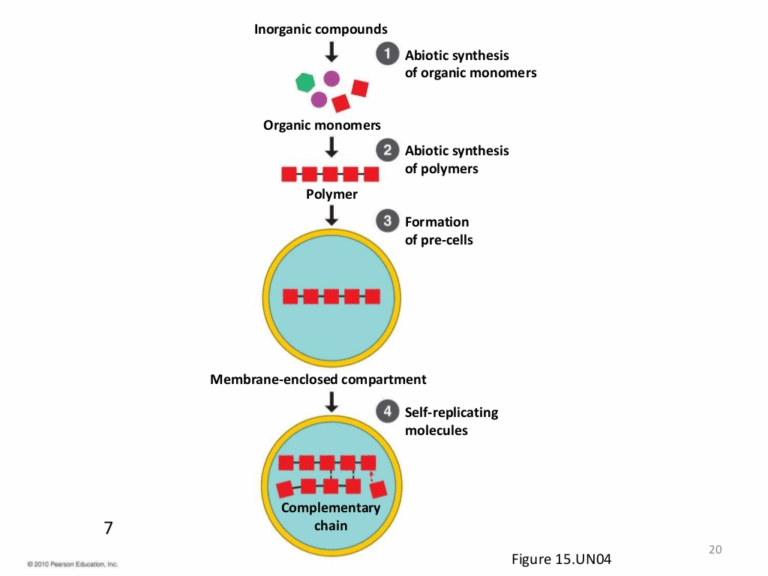
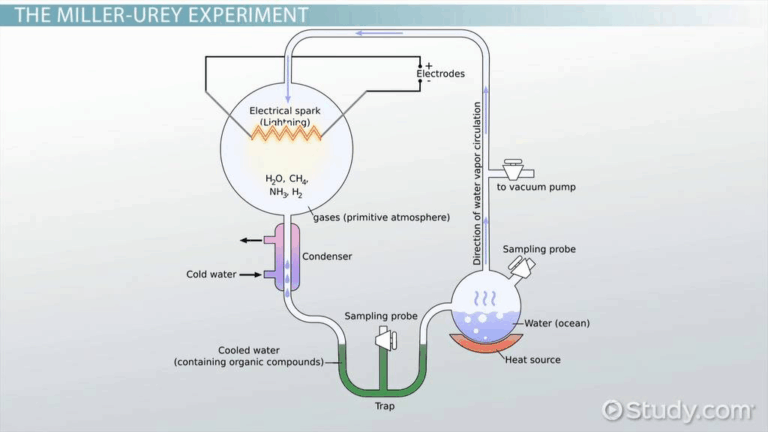
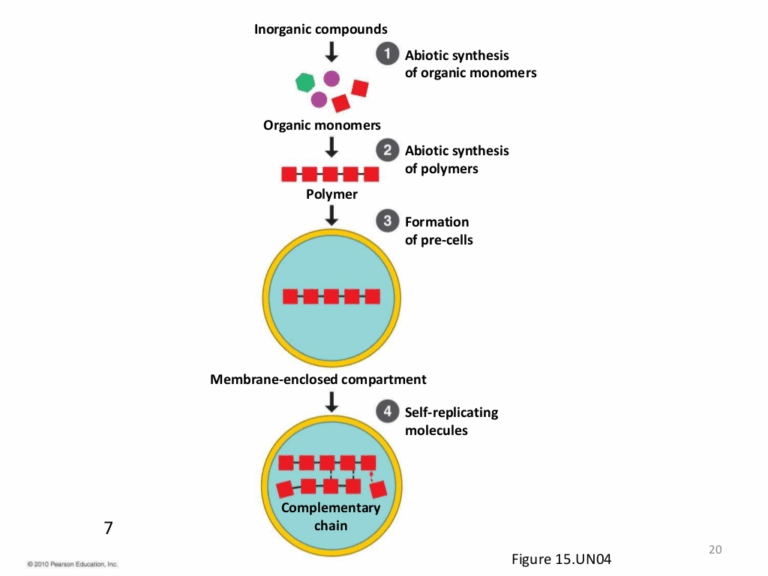
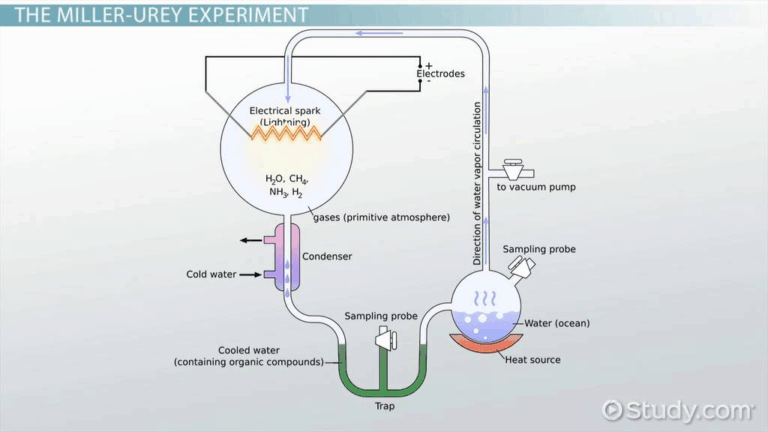
📌 Evolutionary Timescale
- Fossil evidence provides chronological snapshots of past life forms.
- Carbon-14 dating works for relatively recent samples (up to ~60,000 years old).
- Radiometric dating (e.g., uranium-lead dating) measures isotopic decay in older rocks.
- Genome analysis allows estimation of divergence times based on mutation rates.
- The molecular clock assumes mutations occur at a relatively constant rate.
- These methods together allow scientists to approximate when major evolutionary events occurred.
🧬 IA Tips & Guidance: For an IA, consider using simulated mutation rate data to model how molecular clocks estimate divergence times between species.
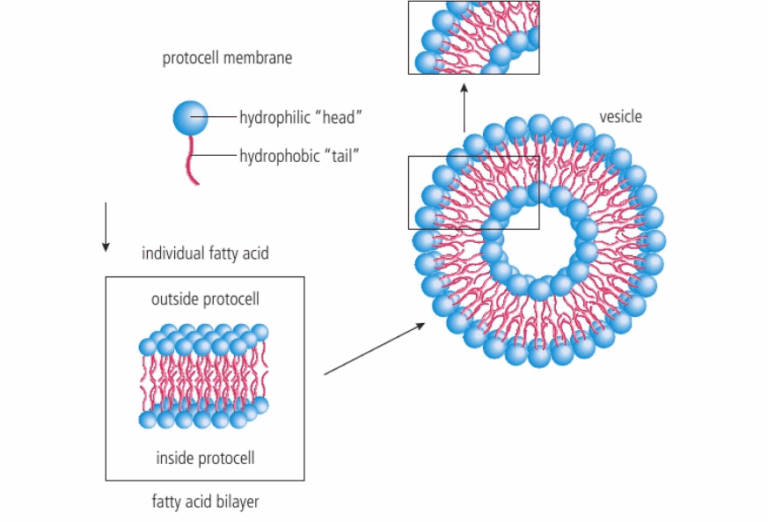
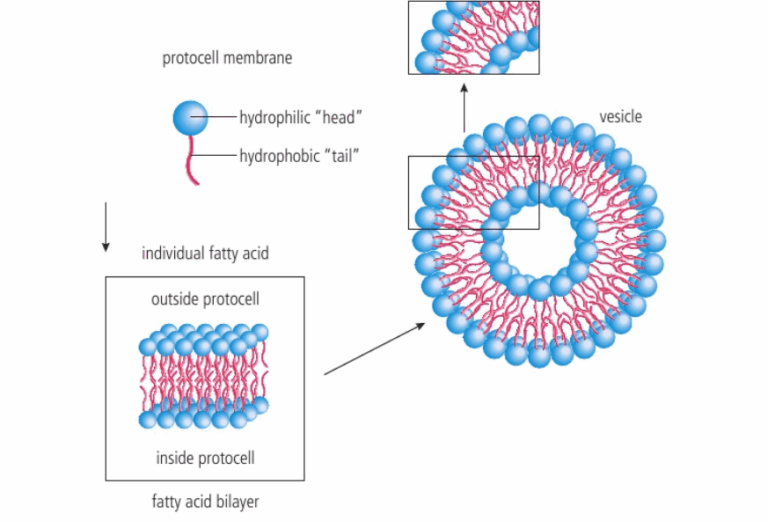
📌 Hydrothermal Vent Hypothesis

- Suggests that LUCA evolved near deep-sea hydrothermal vents.
- Vents provide chemical energy for chemosynthesis, supporting life without sunlight.
- Fossilised haematite tubes in Canada (3.77–4 billion years old) resemble modern vent microbes.
- LUCA may have been an autotrophic extremophile, using CO₂ and H₂ for energy.
- Likely survived high temperatures and lacked oxygen-based metabolism (anaerobic).
- Genetic analysis of modern vent organisms supports a common ancestry with LUCA.
🌐 EE Focus: An EE could explore the biochemical similarities between modern vent extremophiles and the inferred properties of LUCA.
📌 Characteristics of LUCA
- Anaerobic metabolism, using hydrogen as an energy source.
- Ability to convert CO₂ into glucose (autotrophy).
- Nitrogen fixation into ammonia for amino acid synthesis.
- Tolerance for high temperatures (thermophilic).
- Reliance on iron and other minerals abundant in hydrothermal vent environments.
- Basic biochemical pathways similar to those in modern microbes.
❤️ CAS Link: A CAS project could involve developing an interactive timeline of life on Earth, integrating fossil, genetic, and geological evidence.
📌 Challenges in Studying Early Evolution
- Ancient cells did not fossilise, so direct evidence is rare.
- Geological activity has destroyed much of Earth’s earliest rock record.
- Dating methods have margins of error, especially for the oldest samples.
- Molecular clocks require accurate mutation rate assumptions, which may vary between species.
- Fossil and molecular evidence must be interpreted together for reliability.
- Multiple competing hypotheses for life’s origin remain under investigation.
🔍 TOK Perspective: The search for LUCA illustrates the interdisciplinary nature of scientific knowledge — biology, geology, and chemistry must all contribute to construct plausible historical reconstructions, highlighting the role of collaboration in science.
🌍 Real-World Connection:
Understanding LUCA’s traits helps astrobiologists identify possible biosignatures when searching for extraterrestrial life, particularly in extreme environments such as hydrothermal vents on icy moons like Europa.