A1.2.3 – HISTORICAL EXPERIMENTS AND SCIENTIFIC ADVANCES
📌Definition Table
| Term | Definition |
|---|---|
| Transformation | Process where genetic material is transferred from one organism to another, changing its phenotype. |
| X-ray Crystallography | Technique that uses X-ray diffraction to determine molecular structure. |
| Semiconservative Replication | Each new DNA molecule contains one original and one new strand. |
| Model Building | Using physical or computer-based structures to hypothesize biological mechanisms. |
| Radioisotope Labeling | Using radioactive isotopes to trace molecules in experiments. |
📌Introduction
The discovery of DNA’s role and structure was shaped by a series of groundbreaking experiments. From proving DNA as the genetic material to revealing its double-helical structure, these advances laid the foundation of modern molecular biology.
📌 Griffith’s Transformation Experiment (1928)
- Used Streptococcus pneumoniae in mice.
- Smooth (S) strain = virulent; Rough (R) strain = non-virulent.
- Heat-killed S strain + live R strain → mice died.
- Concluded a “transforming principle” transferred virulence.
- Did not identify the molecule responsible.
- First evidence of genetic material transfer.
🧠 Examiner Tip: Always state the organism and strains used when describing historical experiments.
📌 Avery, MacLeod, and McCarty (1944)
- Purified different biomolecules from S strain bacteria.
- Only DNA fraction transformed R strain into virulent form.
- Used DNase, RNase, and protease to confirm DNA’s role.
- Proved DNA was the genetic material in bacteria.
- Paved the way for acceptance of DNA’s central role.
- Still faced skepticism at the time.
🧬 IA Tips & Guidance: Use bacterial transformation to demonstrate uptake of plasmid DNA with visible traits (e.g., fluorescence).
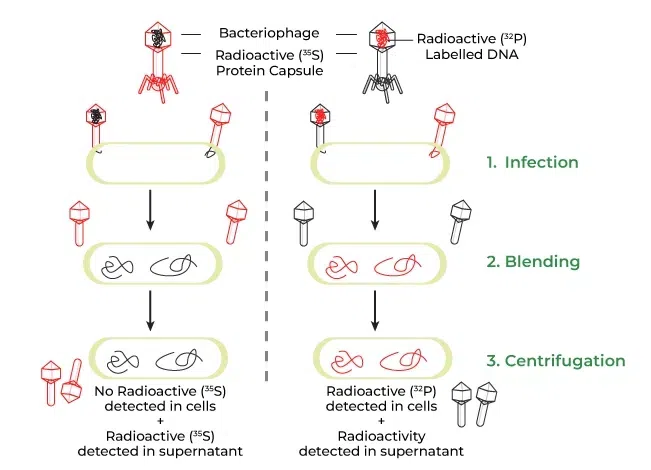
📌 Hershey–Chase Experiment (1952)
- Used bacteriophages labeled with ³²P (DNA) and ³⁵S (protein).
- Only radioactive phosphorus entered bacterial cells.
- Showed DNA, not protein, is the genetic material.
- Used blender and centrifugation to separate phage coats from cells.
- Confirmed Avery’s conclusion in a viral system.
🌐 EE Focus: Compare modern viral genome tracing techniques with Hershey–Chase’s approach.
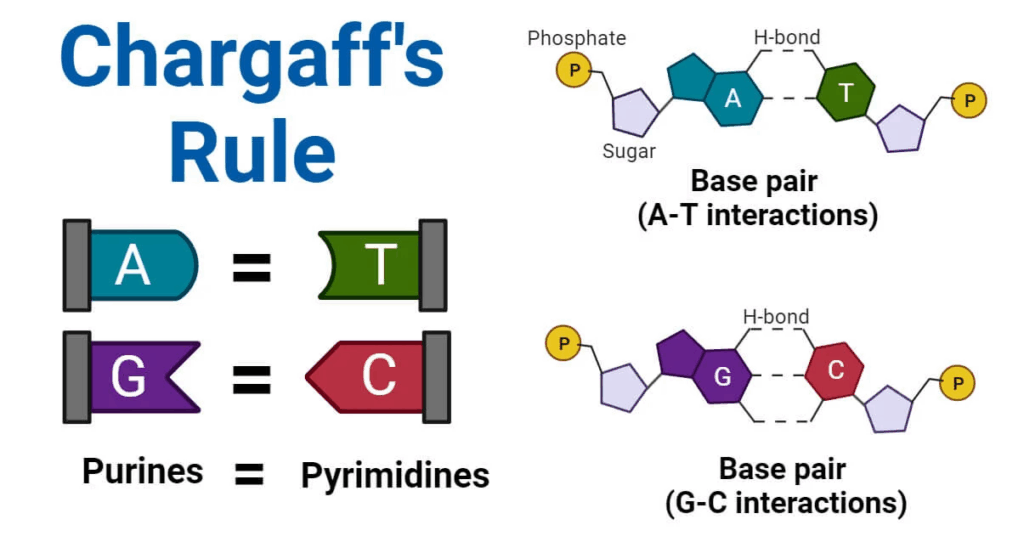
📌 Chargaff’s Rules (1950)
- Analyzed base composition of DNA from various species.
- Found A = T and G = C in molar ratios.
- Ratios vary between species but are constant within one species.
- Provided evidence for complementary base pairing.
- Suggested a structural relationship between bases.
❤️ CAS Link: Create an educational poster showing how Chargaff’s findings led to the base-pairing model.
📌 Franklin and Wilkins (1951–1953)
- Used X-ray diffraction to study DNA fibers.
- Franklin’s Photo 51 showed helical structure with uniform diameter.
- Data indicated two strands and bases stacked inside.
- Helped establish dimensions of the helix.
- Wilkins collaborated with Watson and Crick indirectly.
🌍 Real-World Connection: X-ray crystallography remains vital for studying protein and nucleic acid structures in drug design.
📌 Watson and Crick (1953)
- Built 3D models based on Franklin’s data and Chargaff’s rules.
- Proposed antiparallel double helix with complementary base pairing.
- Explained DNA replication mechanism via strand separation.
- Structure supported semiconservative replication model.
- Their model unified multiple lines of evidence.
🔍 TOK Perspective: Debate exists over ethical issues of credit in scientific discoveries, highlighting the role of collaboration and competition.