D2.2.1 TRANSCRIPTIONAL AND POST-TRANSCRIPTIONAL CONTROL
📌Definition Table
| Term | Definition |
|---|---|
| Transcription factors | Proteins that bind DNA to regulate transcription by promoting or blocking RNA polymerase binding. |
| Promoter | DNA sequence where RNA polymerase and transcription factors initiate transcription. |
| Enhancer | DNA sequence that increases transcription levels when bound by activator proteins. |
| Silencer | DNA sequence that represses transcription when bound by repressor proteins. |
| Post-transcriptional control | Regulation of gene expression after transcription, e.g., RNA splicing, capping, polyadenylation. |
| Alternative splicing | Process in which different exons are combined to produce multiple mRNA variants from one gene. |
📌Introduction
Gene expression in eukaryotes is highly regulated to ensure proteins are produced at the right time, in the right place, and in the correct amounts. Transcriptional control governs whether a gene is transcribed, while post-transcriptional control fine-tunes mRNA after it is produced. These mechanisms underpin cellular differentiation, development, and responses to environmental signals
📌 Transcriptional Regulation
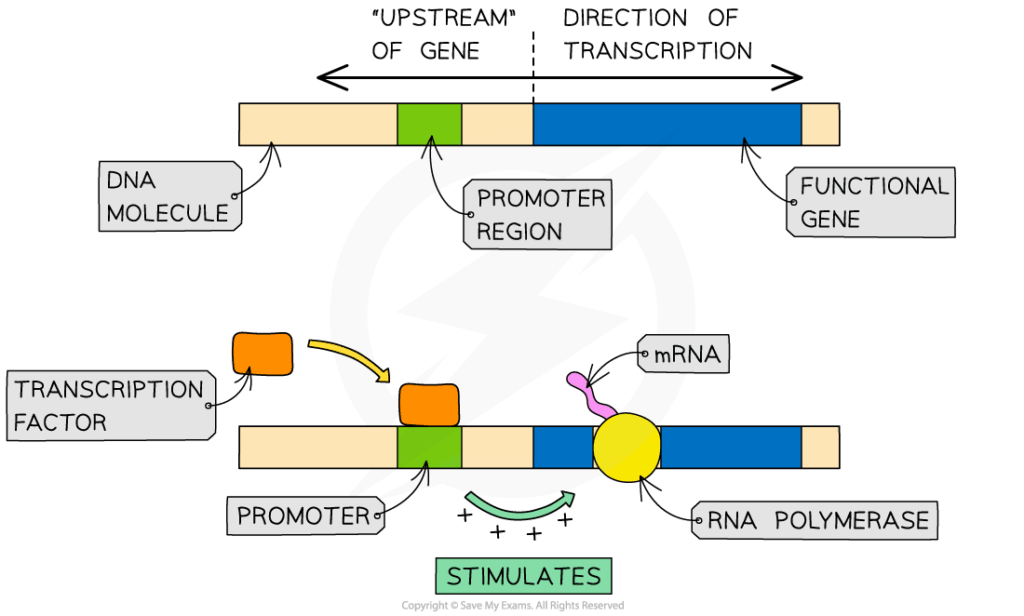
- Promoters: core sequences where RNA polymerase II binds to begin transcription.
- Transcription factors:
- Activators recruit RNA polymerase and stabilise binding.
- Repressors block polymerase or recruit proteins that modify chromatin.
- Enhancers and silencers: act at a distance, looping DNA to contact promoters.
- Specific combinations of transcription factors give each cell type its identity.
- Malfunctions in transcriptional regulation can lead to cancer or developmental disorders.
🧠 Examiner Tip: Don’t just write “transcription factors control transcription.” Name specific mechanisms — e.g., activators at enhancers, repressors at silencers — to gain full credit.
📌 RNA Processing

- 5′ capping: methylated guanine added, protecting mRNA and aiding ribosome binding.
- Poly-A tail: adenine chain added to 3′ end, stabilising transcript and regulating nuclear export.
- RNA splicing: introns removed, exons joined.
- Alternative splicing: allows one gene to code for multiple proteins, increasing proteome diversity.
- mRNA editing can change specific nucleotides, altering protein sequences.
🧬 IA Tips & Guidance: An IA could compare RNA extraction and gel electrophoresis of differently treated cells to show mRNA processing differences.
📌 Nuclear Export and Stability
- Processed mRNA must pass through nuclear pores.
- Export depends on proper capping, tailing, and splicing.
- Cytoplasmic stability of mRNA influences how much protein is made.
- Small RNAs (miRNAs, siRNAs) can degrade specific mRNAs or block translation.
- This regulation ensures quick responses to signals.
🌐 EE Focus: An EE could investigate how alternative splicing produces protein diversity, e.g., in the immune system (antibody variation) or nervous system.
📌 Translation Readiness
- Only properly processed mRNA is translated efficiently.
- Proteins binding untranslated regions (UTRs) regulate ribosome access.
- Iron metabolism regulated by proteins binding ferritin mRNA UTRs.
- This adds another checkpoint for control.
- Demonstrates tight integration of transcription and translation regulation.
❤️ CAS Link: Students could create a workshop or animation to teach juniors how “one gene can make many proteins” through splicing and control mechanisms.
🌍 Real-World Connection: Viruses hijack transcriptional/post-transcriptional machinery. HIV, for example, requires alternative splicing to produce multiple viral proteins.
📌 Integration with Cell Function
- Gene regulation explains cell differentiation in multicellular organisms.
- Errors in splicing can cause diseases like β-thalassemia (faulty haemoglobin production).
- Regulatory mechanisms explain tissue-specific gene expression.
- Understanding control helps develop therapies (antisense RNA, RNAi).
- Highlights complexity of genome usage beyond simple “one gene = one protein.”
🔍 TOK Perspective: Models show neat gene-to-protein pathways, but reality involves layers of regulation. TOK issue: How do simplified models both help and hinder understanding of complex biology?