A3.2.2 – CLADISTICS
📌Definition Table
| Term | Definition |
|---|---|
| Cladistics | A method of classification that groups organisms based on common ancestry and shared derived characteristics (synapomorphies). |
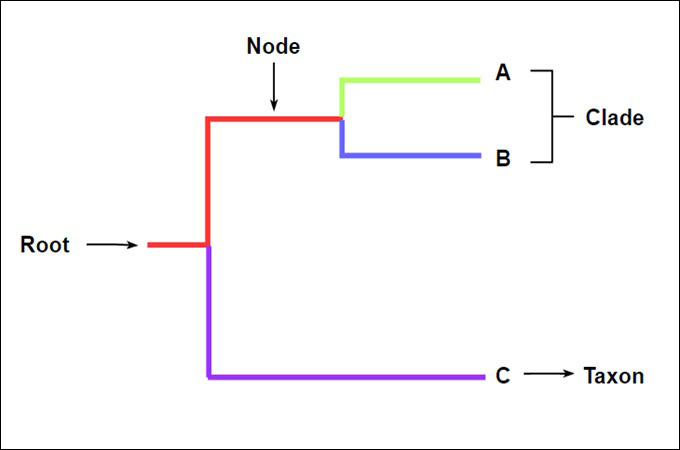
| Clade | A group of organisms consisting of a common ancestor and all its descendants. |
| Synapomorphy | A shared derived trait unique to a clade, indicating common ancestry. |
| Node | A branching point on a cladogram representing the most recent common ancestor of the descendant lineages. |
| Outgroup | A species or group outside the clade being studied, used to infer ancestral traits. |
📌Introduction
Cladistics is a classification approach that focuses on evolutionary relationships rather than overall similarity. It groups organisms into clades, each representing a single branch of the tree of life, defined by shared derived characteristics. Cladograms, the diagrams produced through cladistic analysis, visually represent hypotheses about evolutionary relationships. This method has transformed taxonomy, particularly with the integration of molecular data, making it possible to resolve relationships that morphology alone could not.
📌 Principles of Cladistics
- Groups are based solely on shared derived traits, not ancestral traits.
- All members of a clade share a common ancestor.
- Cladograms are constructed using both morphological and molecular data.
- Branching order represents hypothesised evolutionary pathways.
- Outgroups help determine which traits are ancestral vs derived.
- Cladistics aims for monophyletic groups — including all descendants of a common ancestor.
🧠 Examiner Tip: When drawing cladograms, label each branch with a shared derived characteristic; avoid mixing ancestral traits with synapomorphies.
📌 Construction of Cladograms
- Identify traits and determine which are ancestral vs derived.
- Use an outgroup for reference.
- Assign traits to different branches based on appearance in evolutionary history.
- Arrange branches so that each node represents the emergence of a new trait.
- Molecular data (DNA, RNA, protein sequences) can refine branch placement.
- Branch length may or may not represent evolutionary time — check diagram details.
🧬 IA Tips & Guidance: A possible IA could construct cladograms from genetic sequence data of related plant or animal species and compare them to morphology-based diagrams.
📌 Advantages of Cladistics
- Reflects actual evolutionary history more accurately than traditional systems.
- Allows integration of molecular evidence.
- Avoids polyphyletic and paraphyletic groupings.
- Highlights the evolutionary significance of traits.
- Can be updated easily with new data.
- Provides testable hypotheses about relationships.
🌐 EE Focus: An EE could investigate whether morphological or molecular data produce more accurate cladograms for a specific taxonomic group.
📌 Limitations of Cladistics
- Requires accurate identification of derived vs ancestral traits.
- Convergent evolution can mislead trait-based analysis.
- Incomplete fossil records may limit available data.
- Different data sets (morphological vs molecular) can produce conflicting trees.
- Rapid evolutionary change can obscure relationships.
- Assumes traits evolve in a linear, non-reversible fashion (which may not be true).
❤️ CAS Link: A CAS project could involve teaching younger students how to read and construct cladograms through an interactive workshop.
📌 Molecular Cladistics
- Uses DNA, RNA, or protein sequence comparisons to identify similarities and differences.
- Molecular clocks estimate divergence times between clades.
- Ribosomal RNA sequences are often used due to their slow rate of change.
- Mitochondrial DNA is useful for tracing maternal lineages.
- Data analysis requires bioinformatics tools.
- Can reveal cryptic evolutionary relationships invisible to morphological study.
🔍 TOK Perspective: Cladistics shows how scientific knowledge is shaped by the choice of evidence — morphological and molecular data may lead to different interpretations of evolutionary history.
🌍 Real-World Connection:
Cladistics is used in epidemiology to track virus evolution, in conservation biology to prioritise unique evolutionary lineages, and in agriculture to breed resistant crops.