D1.1.1 STRUCTURE OF DNA AND ENZYMES IN REPLICATION
📌Definition Table
| Term | Definition |
|---|---|
| DNA (Deoxyribonucleic acid) | Double-helical molecule carrying genetic information in base sequences. |
| Nucleotide | Basic unit of DNA, consisting of a phosphate, deoxyribose sugar, and nitrogenous base. |
| Complementary base pairing | Specific hydrogen bonding between bases: A–T (2 bonds), C–G (3 bonds). |
| Helicase | Enzyme that unwinds DNA helix and breaks hydrogen bonds between strands. |
| DNA polymerase III | Main enzyme that synthesizes new DNA strand in 5′→3′ direction. |
| DNA polymerase I | Enzyme that removes RNA primers and replaces them with DNA. |
| Primase | Enzyme that synthesizes short RNA primers to initiate replication. |
| DNA ligase | Enzyme that joins Okazaki fragments by forming sugar–phosphate bonds. |
| Single-stranded binding proteins (SSBs) | Proteins that stabilize unwound DNA strands during replication. |
📌Introduction
DNA replication is essential for genetic continuity in cell division. The double-helical structure, proposed by Watson and Crick, allows complementary base pairing to act as a template for accurate copying. Specialized enzymes coordinate the process, ensuring rapid and precise duplication of billions of nucleotides.
📌 Structure of DNA
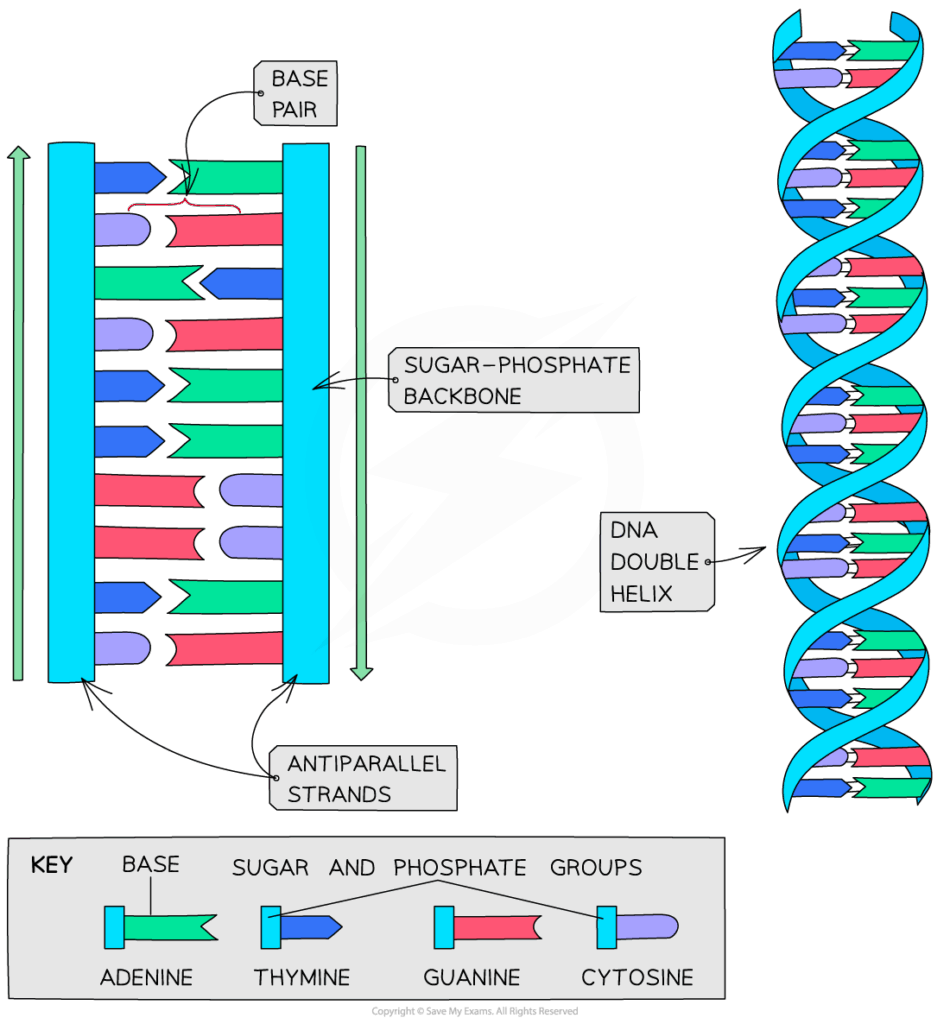
- DNA consists of two antiparallel strands (5′→3′ and 3′→5′) forming a double helix.
- Nucleotides are linked via phosphodiester bonds between sugar and phosphate.
- Hydrogen bonds between complementary bases stabilize the helix: A–T (2 bonds), C–G (3 bonds).
- The antiparallel arrangement means replication requires different strategies for each strand.
🧠 Examiner Tip: In diagrams, always label 5′ and 3′ ends, and show hydrogen bonds as dotted lines between bases.
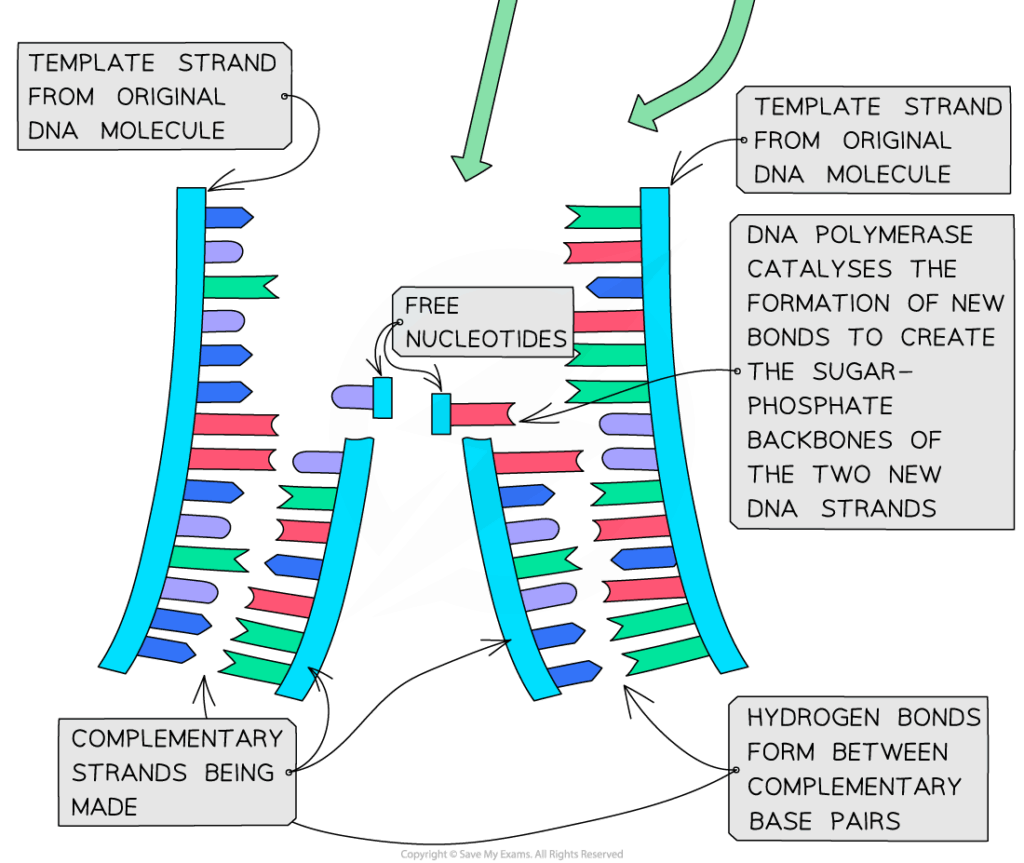
📌 Enzymes in DNA Replication
- Helicase unwinds DNA and breaks hydrogen bonds, creating the replication fork.
- SSBs prevent re-annealing of strands.
- Primase lays down short RNA primers as initiation points.
- DNA polymerase III elongates the new strand by adding nucleotides in the 5′→3′ direction.
- DNA polymerase I removes primers and replaces them with DNA nucleotides.
- DNA ligase joins Okazaki fragments on the lagging strand.
🧬 IA Tips & Guidance: A possible IA could involve modelling DNA replication with molecular kits or computer simulations, emphasizing enzyme roles and directionality.
📌 Importance of Enzymatic Coordination
- Helicase and polymerases must act simultaneously to replicate both strands.
- The coordination of primase, ligase, and proofreading enzymes ensures accuracy.
- Without these enzymes, replication would be error-prone and slow.
🌐 EE Focus: An EE might investigate how inhibitors of DNA replication enzymes (e.g., antibiotics like ciprofloxacin targeting bacterial DNA gyrase) affect cell survival, linking molecular biology to medicine.
📌 Enzymes and Disease Connection
- Malfunction in DNA polymerase proofreading can increase mutation rates.
- Cancer can result from uncontrolled mutations in cell cycle genes.
- Many chemotherapies target enzymes involved in DNA replication.
❤️ CAS Link: Students could design interactive workshops with DNA models, showing how enzymes function in replication, to educate younger students or communities about genetics.
🌍 Real-World Connection:
Antibiotics, antiviral drugs, and chemotherapy often target DNA replication enzymes. For example, AZT (an antiretroviral) inhibits reverse transcriptase in HIV. Understanding replication enzymes underpins medical treatment strategies.
📌 Integration with Genome Stability
- Precise DNA replication preserves genetic identity across cell generations.
- Errors in replication lead to mutations, some of which may drive evolution or disease.
🔍 TOK Perspective: Our knowledge of replication enzymes largely comes from indirect biochemical assays and models. TOK reflection: How do models and analogies (e.g., “unzipping a zipper” for helicase) influence our understanding of complex molecular processes?